Research Tools

Designing Programmable Inducible Promoters for Biosensor Applications
WARF: P170081US02
Inventors: Srivatsan Raman, Aseem Ansari, Xiangyang Liu, Jose Rodriguez-Martinez
The Wisconsin Alumni Research Foundation (WARF) is seeking commercial partners interested in developing de novo methods for designing synthetic inducible promoters. This technology may be broadly applicable across industries where very sensitive detection of small molecules is needed, from medical diagnostics to environmental remediation.
Overview
Biosensors to detect small molecule ligands (e.g., metabolites) have applications in synthetic biology, medical diagnosis, environmental monitoring, bioremediation and bioenergy. Protein-based biosensors are autonomous, self-powered, miniaturizable and programmable macromolecules that function in both in vivo and ex vivo environments. Allosteric transcription factors (aTFs), a family of regulatory proteins found in all kingdoms of life, are widely used as biosensors in synthetic biology.
Natural aTF-promoter pairs, however, may be unsuitable for a wide-range of biosensor applications. Needed are methods of designing synthetic promoters for aTFs that can provide desired induction properties for aTF-based biosensors.
Natural aTF-promoter pairs, however, may be unsuitable for a wide-range of biosensor applications. Needed are methods of designing synthetic promoters for aTFs that can provide desired induction properties for aTF-based biosensors.
The Invention
UW–Madison researchers have developed a method for de novo design of synthetic inducible promoters for transcription factors and other DNA binding proteins such as aTFs with tunable dynamic range behavior and compatibility with virtually any host organism.
The method can include selecting inducible promoters, for example, by converting a constitutive promoter of an organism into an inducible promoter by introducing binding sites near the RNA polymerase binding site. By controlling the access of a transcription factor and the RNA polymerase to the promoter, the dynamic range of the system can be controlled.
The method can include selecting inducible promoters, for example, by converting a constitutive promoter of an organism into an inducible promoter by introducing binding sites near the RNA polymerase binding site. By controlling the access of a transcription factor and the RNA polymerase to the promoter, the dynamic range of the system can be controlled.
Applications
- The method provides for tunable transcription factor-promoter pairs and DNA binding protein-promoter pairs for biosensor uses.
- Medical diagnostics, clean technology, environmental remediation and more
Key Benefits
- Currently, no generalized means of developing aTF-based biosensors exists.
- Most existing methods require screening of fluorescent reporters linked to promoter sequences, which limits versatility.
- The new method provides a means of generating very generic nucleotide sequences that control gene expression through an aTF of interest in a dose-responsive manner.
- Broadly applicable across industries
Stage of Development
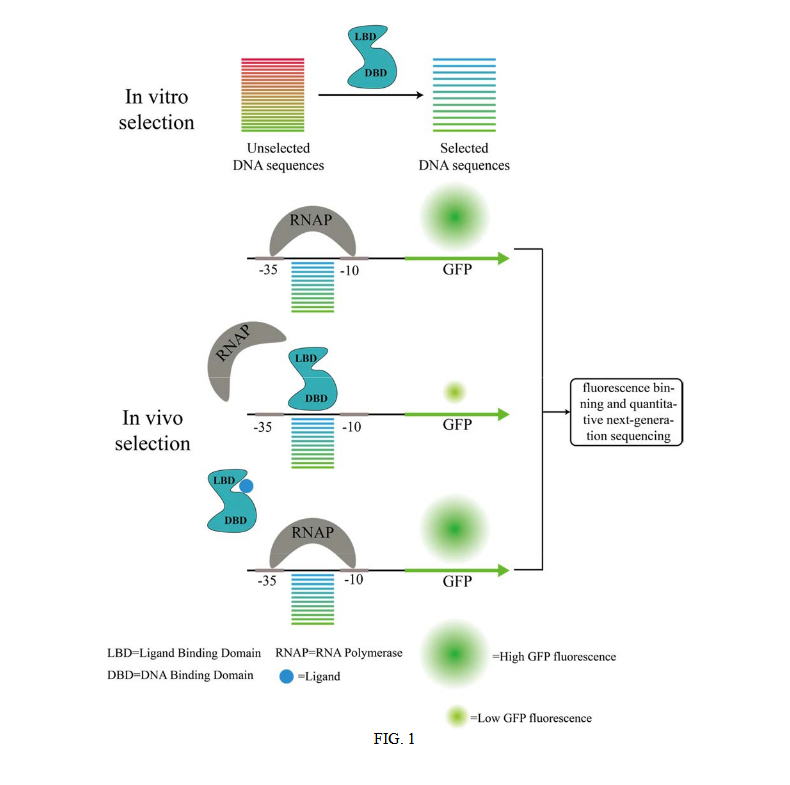
The researchers have developed biosensors using a three-step method: first, in vitro selection is used to identify tens of thousands of DNA sequences with varying affinities to the transcription factor or other DNA binding protein. Then, in vivo transcription of a fluorescent reporter is driven by synthetic promoters containing the in vitro selected transcription factor or other DNA binding sequences to provide a population of polynucleotides that repress or induce production of the reporter protein.
Finally, the induction properties of the promoters (tens of thousands) are simultaneously characterized by fluorescence binning and quantitative next-generation sequencing to enable selection of synthetic promoters having a specified induction and/or repression for the transcription factor or other DNA binding protein and the inducer molecule. (See Figure 1 from the patent application below.)
Finally, the induction properties of the promoters (tens of thousands) are simultaneously characterized by fluorescence binning and quantitative next-generation sequencing to enable selection of synthetic promoters having a specified induction and/or repression for the transcription factor or other DNA binding protein and the inducer molecule. (See Figure 1 from the patent application below.)
Additional Information
For More Information About the Inventors
Tech Fields
For current licensing status, please contact Jennifer Gottwald at [javascript protected email address] or 608-960-9854
Figures